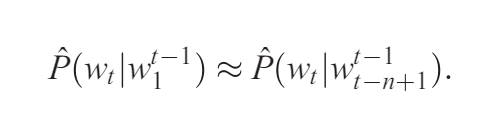total number of names: 102449
min length word: 2 max length word: 15
shortest_names: ['ii', 'ja', 'vi', 'od', 'kd', 'ax', 'jd', 'jp', 'st', 'sa', 'rc', 'jt', 'xi', 'ju', 'zy', 'mi', 'kj', 'cj', 'ho', 'se', 'io', 'ge', 'eh', 'jw', 'un', 'kc', 'no', 'an', 'mr', 'va', 'oz', 'du', 'ji', 'ah', 'tr', 'mc', 'si', 'zi', 'ld', 'go', 'pj', 'la', 'qi', 'jm', 'or', 'bj', 'sy', 'lu', 'ao', 'zo', 'su', 'ed', 'xu', 'za', 'ra', 'bb', 'na', 'ry', 'ki', 'pa', 'gy', 'md', 'vu', 'fu', 'ti', 'lj', 'jo', 'ad', 'ej', 'di', 'jl', 'my', 'ku', 'mu', 'lc', 'vy', 'te', 'ar', 'aj', 'ze', 'rb', 'ly', 'jc', 'el', 'so', 'ya', 'ma', 'gi', 'ia', 'yu', 'po', 'li', 'ac', 'lb', 'sj', 'tu', 'ke', 'fe', 'ro', 'kt', 'dj', 'al', 'eb', 'wa', 'mj', 'ab', 'oh', 'rj', 'tc', 'je', 'hy', 'lg', 'yi', 'om', 'yy', 'oc', 'ty', 'me', 'ko', 'av', 'ny', 'ng', 'yo', 'ai', 'jb', 'ka', 'jj', 'ru', 'ea', 'ni', 'ky', 'da', 'rd', 'de', 'le', 'bo', 'do', 'ta', 'rl', 'jr', 'ye', 'in', 'mo', 'ok', 'wc', 'hu', 'wm', 'ha', 'bg', 'ba', 'be', 'lo', 'cy', 'tj', 'en']
longest_names: ['laurenelizabeth', 'ryanchristopher', 'christianjoseph', 'sophiaelizabeth', 'mariadelosangel', 'michaelchristop', 'ashleyelizabeth', 'johnchristopher', 'muhammadibrahim', 'jordanalexander', 'joshuaalexander', 'christophermich', 'christopherpaul', 'christianmichae', 'christianalexan', 'jonathanmichael', 'christiandaniel', 'davidchristophe', 'gabrielalexande', 'christopherdavi', 'mariadelrosario', 'christopherjose', 'christopherjohn', 'jordanchristoph', 'markchristopher', 'seanchristopher', 'christopheranth', 'kevinchristophe', 'christopherjame', 'jaydenalexander', 'christiananthon', 'christopherryan', 'muhammadmustafa', 'franciscojavier', 'hannahelizabeth', 'christianjoshua', 'matthewalexande']
random sample of 5 names: ['kinzi', 'jennifermarie', 'brayce', 'kentrevious', 'adoniram']